about.
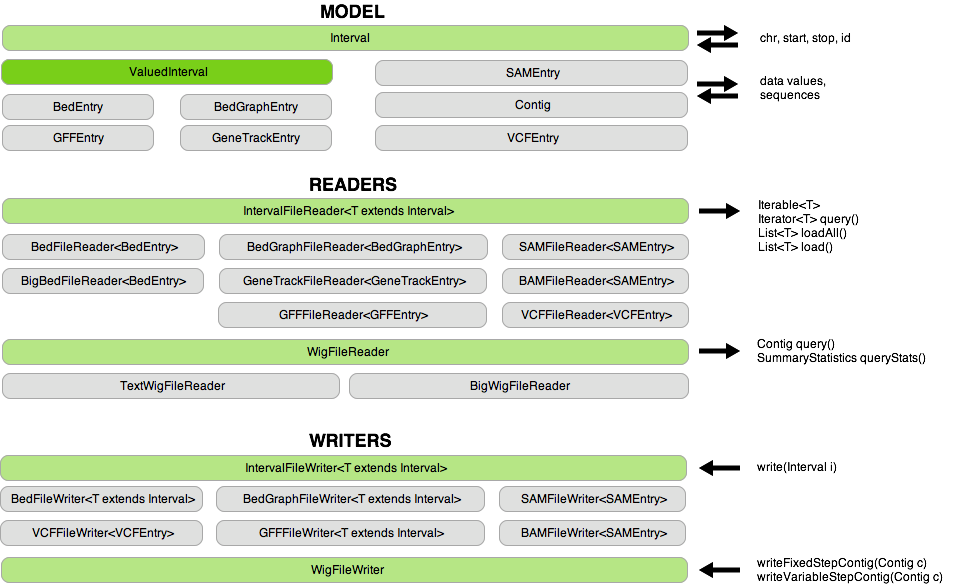
This library provides a simple and performant API for reading and writing many genomic file formats in a consistent fashion. File formats currently supported include: Bed, BigBed, BedGraph, GFF, GeneTrack, SAM, BAM, VCF, Wiggle, BigWig, and Tabix.
The design seeks to accomplish two major tasks:
- Allow many different formats to be accessed and manipulated in a consistent way without needing to worry about parsing individual file types. For example, all line-based interval formats (Bed, BedGraph, BigBed, GFF, GeneTrack, SAM, BAM, VCF) inherit from the Interval class so that applications may be agnostic to the specific format of input files. In a similar fashion, text Wiggle files and BigWig files share the same interface and may be used interchangeably.
- Make random-access to information in large genomic datasets efficient by using indexing schemes. In this way, your tools can randomly pull information from specific regions of the genome without the major performance cost incurred by seeking through ASCII text files line-by-line. Line-based interval files are indexed as needed using a pure-Java reimplementation of Tabix. Built-in indexes are used for BAM, BigWig, and BigBed files. ASCII Wiggle files are indexed using a custom implementation.
With all readers, data is buffered and lazy-parsed to minimize memory requirements and maximize disk performance.
Writers are also available to create line-based interval files and Wiggle files.
usage.
For code and usage examples, see GitHub. Some examples of full script implementations are available in the java-genomics-toolkit.
api.
JavaDocs are available online and may also be generated with the ant task "javadoc".
requirements.
java-genomics-io requires Java 7, available at oracle.com. In addition, it requires several external libraries that are included with the distribution. Make sure to include these jar files in the classpath if you use java-genomics-io in your own application.
download.
The recommended way to obtain the library is to check out the source code from GitHub and build it using the provided Ant build script (simply call "ant").
java-genomics-toolkit.
For some examples of tools built with this library, see the java-genomics-toolkit, which provides a suite of tools for processing, analyzing, and visualizing next-generation sequencing data.
licensing.info.
java-genomics-io is distributed under the GNU General Public License v3. See the included license.txt for more details.
java-genomics-io was created by Timothy Palpant for work in the Lieb laboratory at UNC Chapel Hill.
java-genomics-io utilizes multiple external libraries including:
- BigWig/BigBed Reader (by the Broad Institute) to read BigWig/BigBed files
- Picard (SAM-JDK) for reading SAM/BAM files
- log4j for logging
- commons-lang for String and Array manipulation
- commons-math for mathematics
contact.me.
Please contact me at tim [at] palpant.us with bugs, questions, comments, or suggestions.